New tool opens up old data and discovers novel links between exercise and health
A free online tool that lets anyone discover how different groups of people respond to exercise, has been launched by a team of international researchers including CBMR Executive Director Juleen Zierath. MetaMEx pools data from 66 studies that examined the impact of exercise on gene behaviour in muscles and holds enormous potential for the future development of personalised medicine and exercise therapy, say the researchers from Karolinska Institutet, the University of Copenhagen, and the Institute for Health and Sport, Victoria University.

We know that exercise can play a powerful role in preventing or treating metabolic diseases, like obesity and diabetes. But scientists are still not sure how factors such as age, gender and genetics have an impact on the effectiveness of different types of exercise, such as resistance, aerobic or high-intensity interval training (HIIT). If we did, it could unlock the therapeutic potential of personalised training programs and medications.
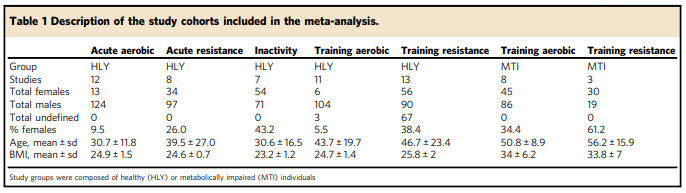
Many studies have already examined how exercise and inactivity change the behaviour of genes in muscle cells. While these studies often produce new insights, they do not always provide a clear direction for scientists who are interested in discovering the most effective forms of exercise for different groups of people. This is because the studies were small and often included a limited cross-section of the population, and we already know that people can respond very differently to different types of exercise.
Researchers from Karolinska Institutet and the University of Copenhagen realised that we might already have the data we need to start answering these questions. In an article in Nature Communications, they explain how they collected publicly-available data from 66 different studies into an online meta-analysis tool called MetaMEx.
“We collected all gene expression data and clinical data available so that we could group studies by age, sex, protocol, and so on. This creates a unique resource of exercise studies, all available in one place,” explains Nicolas Pillon, assistant professor at Karolinska Institutet and the article’s lead author.
Anyone can now dig into the data
The tool exploits the untapped potential of these studies and gives users insight into how hundreds of different genes behave after exercise or during inactivity. Users can filter the data based on the subject’s age, sex, level of fitness and health status, which provides the ability to easily compare how these groups differ.
Nicolas Pillon explains how technological advancements have made a tool like Meta Mex possible:
“Big data analyses like RNA sequencing are now widely used. These techniques allow a deep understanding of biology, but require advanced bioinformatic skills. Most data are uploaded in repositories and freely available, but are of little use to people without the adequate programming knowledge. With our MetaMEx online tool, anyone can interrogate all published exercise databases in a few clicks.”
The researchers also demonstrated MetaMEx’s potential. Diving into the data, they discovered that the NR4A3 gene was one of the genes that responded most strongly to both exercise and inactivity. By carrying out their own laboratory tests on muscle cells, the researchers confirmed that their observation in MetaMEx was actually the case.
The researchers argue that one strength of the tool is that it can be continually updated with new data, which only strengthens its predictive power. However, for the tool to reach its full potential, studies need to provide more detailed information about how and when the biological samples are taken from the subjects.
Co-author Juleen Zierath, Professor at both the University of Copenhagen and Karolinska Institutet, also points out that the studies lack diversity.
“One interesting aspect of our analysis is to point at gaps in knowledge and understudied populations. For instance, there are fewer exercise studies performed in women, so we don’t get a full picture of how exercise affects females’ genes.”
A step toward personalised exercise interventions
The tool allows researchers to compare how different types of people respond to different types of exercise. By finding ways that groups respond differently, it could give researchers new lines of enquiry to pursue in developing treatments that work better for people depending on their unique circumstance.
“Studies in homogeneous cohorts are useful but difficult to extrapolate to the general population,” explains Juleen Zierath, adding:
“We know that not everyone responds the same to exercise. Understanding the diversity of exercise responses will improve how we design specific exercise regimes for different populations to maximise the benefits of physical activity. Our study includes more than a thousand individuals of various age, sex and disease and has the potential to identify selective responses that could be targeted to improve healthspan.”
Read the full article in Nature Communications here: 'Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity'
