Research Program 1
Human Variation and Functional Genomics
The goal of Research Program 1, Human Variation and Functional Genomics, is to understand the biological mechanisms that regulate cardiometabolic health and disease through studying the intricate interplay between the human genome, its molecular derivatives and the environment.
Scope
The Program will leverage high-resolution discovery, variant-to-function translation, and subtyping approaches to generate knowledge, catalyzing the transition from one-size-fits-all prevention and treatment strategies to more personalized care.
Program 1 rests on the fundamental assumption that variation in the human genome and its downstream molecular derivatives drive cardiometabolic disease. Identifying this variation is not only instrumental in better understanding the underlying mechanisms, but also foundational to provide molecular insights enabling more precise diagnoses, prognoses and treatment options.
Program 1 considers the following thematic areas out of scope: variant-to-function work in the pancreas, and general mapping and exploration of the microbiome.
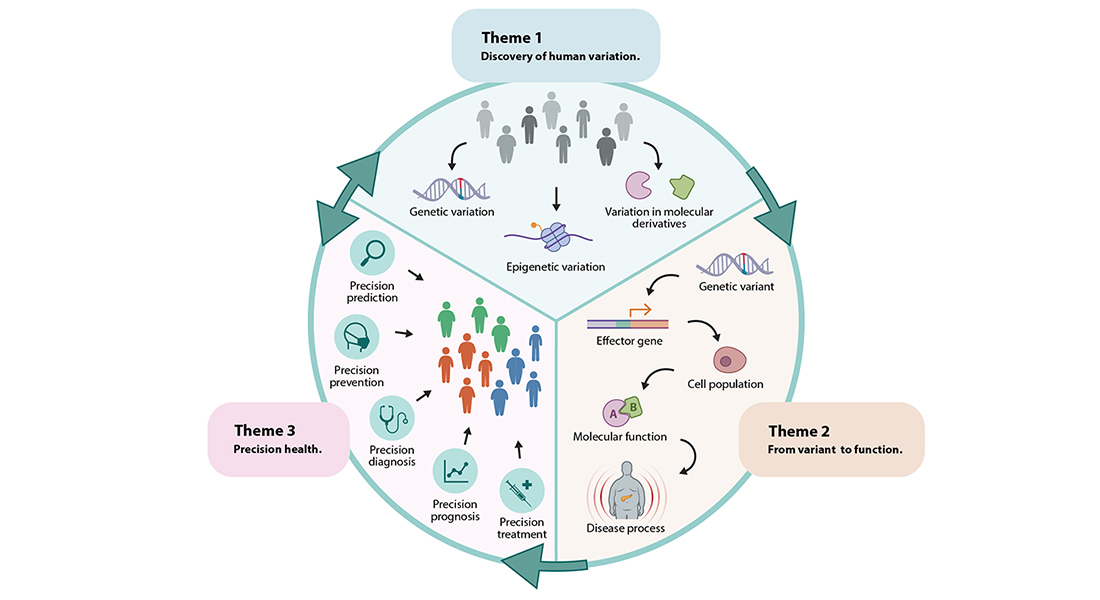
To close the knowledge gaps between cardiometabolic trait-associated molecular variants, etiologic molecular pathways and clinical variation observed in individuals with cardiometabolic conditions, Program 1 will focus on three key themes. In its first theme (Discovery of Human Variation), the Program focuses on discovery of genetic and molecular variation that contributes to cardiometabolic conditions. Large-scale agnostic discovery analyses to identify the contribution of variation in the genome and its molecular derivatives to cardiometabolic disease will be applied in well-phenotyped Danish and international cohorts. In its second theme (Variant to Function), Program 1 applies variant-to-function, single-cell and machine-learning approaches in relevant cell populations to map the genes and molecular processes that underlie cardiometabolic disease. In its final theme (Precision Health), Program 1 aims to dissect the heterogeneity in cardiometabolic disease by identifying molecularly-defined homogeneous subtypes.
Despite the enormous progress in genetic association and variant-to-function approaches, only the first layers of the underlying mechanisms have been revealed. Program 1 aims to target the deeper layers of the biology, implementing advanced approaches that improve precision at the “exposure” side and “outcome” side of the association equation. Discovery analyses will not only identify variation in the human genome (including rare and common variants, chromatin modifications), but also in its molecular derivatives (including RNAs, proteins, metabolites, peptides and hormones), and in environmentally regulated factors (including the microbiome and exogeneous metabolites) that affect individuals’ risk of developing cardiometabolic disease.
We will use Mendelian randomization approaches to identify new biomarkers that are causally related to cardiometabolic outcomes. In terms of the outcomes, discovery analyses will move beyond the traditional “crude” outcomes to more precise phenotypes (including adipose tissue morphology, oral glucose tolerance tests, postprandial glucose levels) and multi-trait analyses.
Despite the discovery of numerous genetic associations to cardiometabolic traits and diseases, little is known about the molecular mechanisms through which they exert their effect. To fill this gap, Program 1 aims to connect genetic variants with cell population-specific effector genes and multi-omic molecular and imaging-based readouts. Focusing on adipocytes, myocytes and hypothalamic cells, Program 1 will leverage human tissue samples to identify cell population-resolved chromatin accessibility and transcriptional states that correlate with cardiometabolic conditions. In addition, the Program will apply techniques from the Genetic Perturbation and Single-Cell Omics Platforms on human induced pluripotent stem cells (hiPSC) and cell villages derived from well-phenotyped donors, to understand and validate the mechanisms with which genetic variants impact expression of effector genes and activity of gene-regulatory networks. Machine learning techniques will be used to predict specific molecular processes impacted by a given set of genetic variants.
The science of this theme is expected to highlight subsets of genetic variants acting through relevant cell populations (for example waist-to-hip ratio-associated genetic variants exerting their effects through adipocytes), candidate effector genes mediating the effect of genetic variants, and molecular processes on which these effector genes coalesce. Translation of only a few dozen of the GWAS-identified loci could already tremendously expand our insights into the biology of cardiometabolic disease, and possibly reveal new therapeutic targets. It would also take us a little closer to the ability to move away from a ‘one-size-fits-all’ strategy, and towards truly precision medicine for cardiometabolic diseases.
Cardiometabolic conditions are complex, multifactorial diseases that result from an intricate interplay between genetic and environmental factors. Despite their complexity, these diseases are typically defined by one simple metric, e.g., BMI≥30 kg/m2 for obesity, or fasting glucose≥7mM for type 2 diabetes. It is therefore no surprise that these conditions comprise a heterogenous group of individuals. To account for this heterogeneity, Program 1 aims to subclassify these complex conditions into smaller, more homogeneous subtypes. The assumption is that stratification of the population in subgroups forms the basis of an improved and more precise diagnosis (precision diagnosis), which in turn forms the foundation for tailored, subgroup-specific treatment and prevention strategies (precision treatment and precision prevention). Disease subgroups will also improve prognosis (precision diagnosis) and facilitate prediction (precision prediction).
So far, subclassifications have been predominantly performed based on clinical features in individuals that have obesity, diabetes or cardiometabolic disease (i.e., phenotypic subclassification). Within subgroups of phenotypically distinct individuals, Program 1 in this research theme, aims to identify genetic and molecular markers that enable the Program to draw new insights in the etiology of its subtypes, which in turn may enable more precise prognoses and treatment options, and, in contrast to clinical biomarkers-based approaches, more timely prevention.


