Fluxomics
Metabolomics can provide indications of an organism’s physiological status at a given time under given conditions. This can also be seen as a snapshot. However, snapshots of the metabolites’ pool sizes have limitations, and can therefore can be complemented by determination of metabolic fluxes and dynamics analyses. This will provide additional information of an organism’s metabolic responses to genetic or environmental changes. A well-established method for flux analysis is feeding with stable isotope labelled tracers such 13C, 15N or 2H. Feeding of 13C-labelled substrate such as 13C-glucose to a cell culture/line is well established, and the 13C compatible with mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR) techniques, and therefore degree of labelling is easily measured.
Additionally, isotopic labeling experiments help us understand which substrates a given metabolite is derived from and can provide crucial information about pathway activity. In the initial state, after correcting for the natural abundance of isotopes, the isotope mass distribution ratio of each metabolite is zero. After growing the system on an isotopically labeled substrate for a while, labeled atoms will be incorporated into downstream metabolites (see figure below). If the mass isotope distribution indicates that labeled atoms are present, then it can be argued that the metabolite is derived from the labeled substrate. Once the mass distribution of the metabolites does not change over time anymore, the system has reached isotopic steady-state, a prerequisite for steady-state Metabolic Flux Analysis.
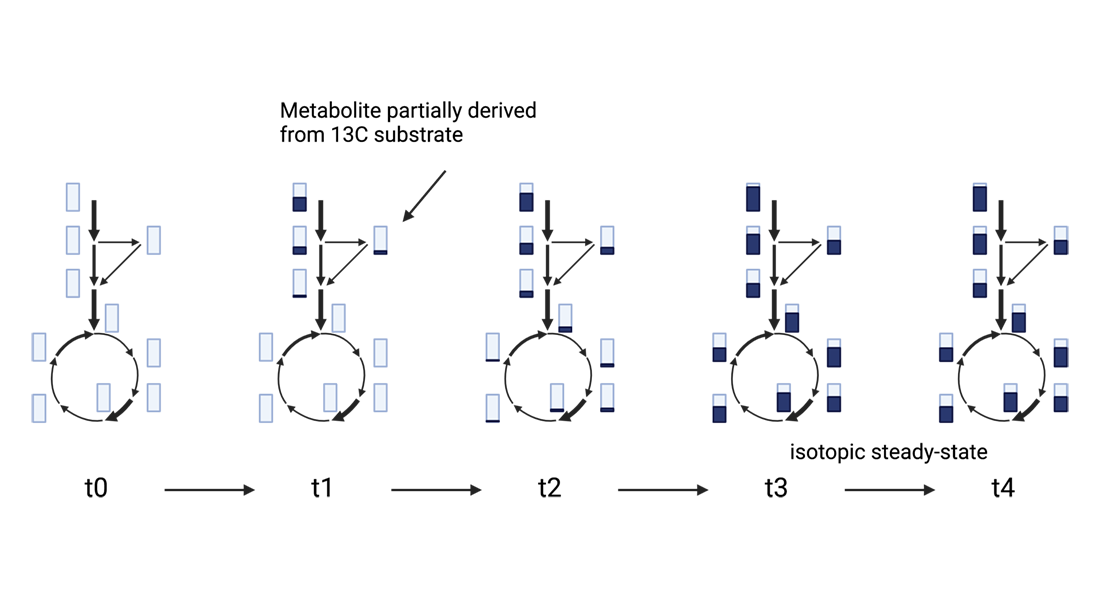
Metabolomics Services
Interested in a service? Fill out the Service Request Form below
