Genomic Physiology and Translation in the Hansen Group
The Hansen Group investigates the human genome and the human global microbiome to characterize the impact of these genomes on health and disease and to identify interactions between the host and the microbiome. The findings can lead to potent translational strategies for prevention and treatment of metabolic disease.
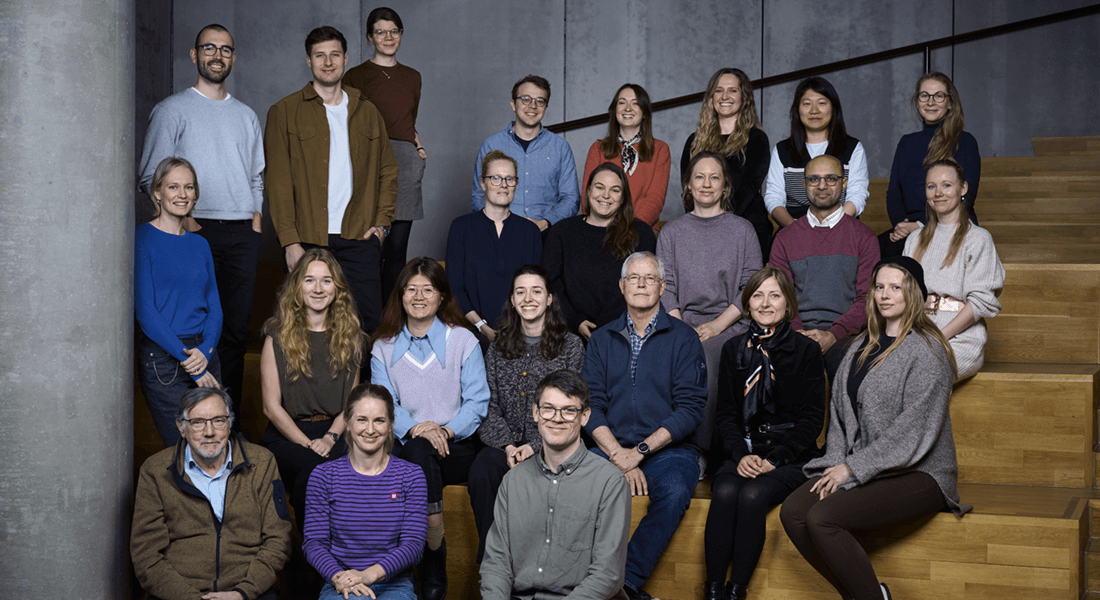
2nd row (left to right): Roman Thielemann, Thomas Gade Koefoed, Mette Andersen Lepola, Anne Cathrine Baun Thuesen, Lise Ryborg, Sufyan Suleman, Kimmie Vestergaard Sørensen
3rd row (left to right): Stina Ramne, Shanshan He, Marina Kanellou, Torben Hansen, Tina Hvidtfeldt Lorentzen, Maria Heinesen
Bottom row (left to right): Thorkild I. A. Sørensen, Pauline Kromann Reim, Rasmus Tanderup Jensen
In the molecular understanding of metabolic diseases, a major gap exists between basic genetic and microbiome discoveries and their impact on physiology and the potential for clinical translation. The Hansen Group aims to bridge this gap by connecting genomic discoveries and epidemiology, with physiology and clinical application.
To study the role of selected genetic variants in human metabolism, we perform physiology and intervention studies based on recruit-by-genotype principles. We also investigate families and populations with extreme metabolic phenotypes and perform physiology and intervention studies.
Finally, we investigate targeted clinical management of carriers of selected high-impact variations in the human genome.
”Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes”
Published in Nature Genetics in 2018, this study identifies a variant in ADCY3 (encoding adenylate cyclase 3) associated with markedly increased risk of obesity and type 2 diabetes in the Greenlandic population and observes an enrichment of rare ADCY3 loss-of-function variants among individuals with type 2 diabetes in trans-ancestry cohorts.
"Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes"
Published in Nature Genetics in 2019, this study demonstrates that genetic variation in hubs impacts insulin secretion heritability, and show that hub annotations can be used for polygenic scores that predict T2D risk driven by islet regulatory variants. Human islet 3D chromatin architecture, therefore, provides a framework for interpretation of T2D genome-wide association study (GWAS) signals.
"Environmental Spread of Microbes Impacts the Development of Metabolic Phenotypes in Mice Transplanted With Microbial Communities From Humans"
Published in ISME Journal in 2018, this study shows that Microbiota transplantation to germ-free animals is a powerful method to study involvement of gut microbes in the aetiology of metabolic syndrome. Owing to large interpersonal variability in gut microbiota, studies with broad coverage of donors are needed to elucidate the establishment of human-derived microbiotas in mice, factors affecting this process and resulting impact on metabolic health. Our findings highlight the impact of donor variability and reveal that inter-individual spread of microbes contributes to the development of metabolic traits. This is of major importance for design of animal studies, and indicates that environmental transfer of microbes between individuals may affect host metabolic traits.
Microbiome in liver and metabolic diseases
The microbiome, i.e. the vast range of bacteria, viruses and fungi colonizing the body, plays important roles within nutrient degradation and protection from infection. In addition, the microbiome composition has been associated with a range of metabolic diseases, including obesity and diabetes. In the Hansen group, we investigate how the gut and oral microbiome impacts metabolic health, both in general and focusing on specific diseases, such as diabetes and fatty liver disease. In addition to the microbiome shotgun metagenomic sequencing, we use data from whole-genome SNP arrays to assess the relative roles of host genetics and environment on the human microbiome. We are also collaborating with experts in metabolomics, proteomics, and other big data fields, as well as computer scientists working towards integration across ‘omics data.
Metabolic disease
In the ongoing obesity epidemic, metabolic diseases are widespread in the general public. In the Hansen group we work with a number of large population cohorts, including “Diet, Cancer and Health – Next Generations” and Inter99, which are both studies involving families or longitudinal assessments. These projects are aimed at understanding early pathogenesis of metabolic disease.
Liver disease
The Hansen group is currently involved in several projects (MicrobLiver, GALAXY (http://www.livergalaxy.eu/), MICROB-PREDICT (https://www.microb-predict.eu/) investigating the pathogenesis of liver diseases. These include alcoholic and non-alcoholic liver disease, from early stage development in children with obesity (TARGET cohort) through population monitoring (Inter99 study), outpatient clinics (GALAXY, SIPHON), and a number of clinical intervention studies (weight loss or training, antibiotic or probiotic treatment, gastric bypass surgery, and transjugular intrahepatic portosystemic shunt (TIPS) surgery). The genetics of liver disease, and its relationship with metabolic diseases are being investigated using methods including polygenic risk scores (PRS) and Mendelian Randomization, along with genome-wide association studies (GWAS) for microbiome, metabolomic and liver phenotypes.
Application of human genetics for classification and treatment of diabetes and obesity
Common variants has an established effect on the development of several diseases such as diabetes and obesity - either as single variants or when combining variants into polygenic risk scores. Such risk scores can facilitate the sub-classification of patients, which may support precision therapy. Neither single common variants nor polygenic risk scores are yet clinical applicable in type 2 diabetes. In the Hansen group, we are involved in projects examining the effect of single common variants or polygenic risk scores on physiology including beta-cell function and incretin regulation. These studies are performed using data from genome-wide SNP analyses in cohorts, which are deeply phenotyped such as the population-based Inter99 study.
Rare variants can also be disease causing including certain variants in genes such as GCK, HNF1A, HNF4A causing monogenic diabetes and rare variants in genes such as LEP, LEPR, MC4R involved in the development of monogenic obesity. For several of these genes, targeted treatment based on a genetic diagnosis is available. Despite the possibility to apply precision medicine based on genetic diagnostics, a number of patients do not undergo genetic screening and are not offered the optimal treatment. In the Hansen group, we are involved in projects investigating the prevalence of monogenic diabetes and obesity in different cohorts, developing methods for estimating the pathogenicity of specific variants (PRISM) and implementing the application of precision medicine in diabetes (TRANSLATE). We are also involved in studies aiming at the identification of novel monogenic forms of diabetes and obesity. We are furthermore involved in the understanding of the interaction between common and rare variants as well as the functional influence on human genetic variants of factors such as the composition of the gut microbiome.
These investigations are being performed using targeted resequencing, exome sequencing and whole-genome sequencing in a nation-wide cohort of newly diagnosed type 2 diabetes patients (DD2), in patients with diabetes recruited from Danish out-patient clinics, in women with gestational diabetes as well as obese children of different ethnicities.
Population isolates: application in genetic association studies
The genetic predisposition to a range of metabolic traits, including diabetes and obesity, is studied intensively to shed light on the biological mechanisms underlying these conditions, and eventually to improve prediction and prevention of disease. Small and historically isolated populations as well as consanguine families are ideal for genetic association studies, due to their unique demographic and family history, which allows for high impact variants to be more common and thus easier to identify and characterize.
In the Hansen group, we are involved in a range of project aiming at identifying novel genetic variants associated with metabolic conditions in the Greenlandic study cohorts IHIT, B99, and B2018 collected as part of the Greenlandic population surveys, and in consanguineous families from the Middle East, northern Africa, and the United Arabic Emirates . Genetic loci identified so far in the Greenlandic cohorts, include TBC1D4, associated with increased risk of type 2 diabetes, and ADCY3 associated with increased risk of obesity. The increased power obtained by studying an isolated population, further allows for analyses applying other genetic models than the additive model most commonly used in genetic association studies. Hence, in the Hansen group, we have identified novel type 2 diabetes risk loci by applying a recessive genetic model.
Besides access to a broad range of phenotypes, and genotype data generated by chip-genotyping and whole genome sequencing, the Hansen group also work to generate additional levels of data, including RNA expression, proteomics, metabolomics, and gut-microbiome data, in order to generate a study sample, which encompasses multiple levels of integratable data.